Friederike Ehrhart
CC-BY 4.0
This blog post shows a few examples of how to use the eNanoMapper database search function to find specific data:
1. how to find data about TiO2 nanoparticles?
2. how to find data of carcinogenicity experiments with nanoparticles?
3. how can we merge data from different assays?
4. which nanomaterials in the database contain PEG?
5. what happens if there is nothing in the database about my material?
1. How to find data about TiO2 nanoparticles?
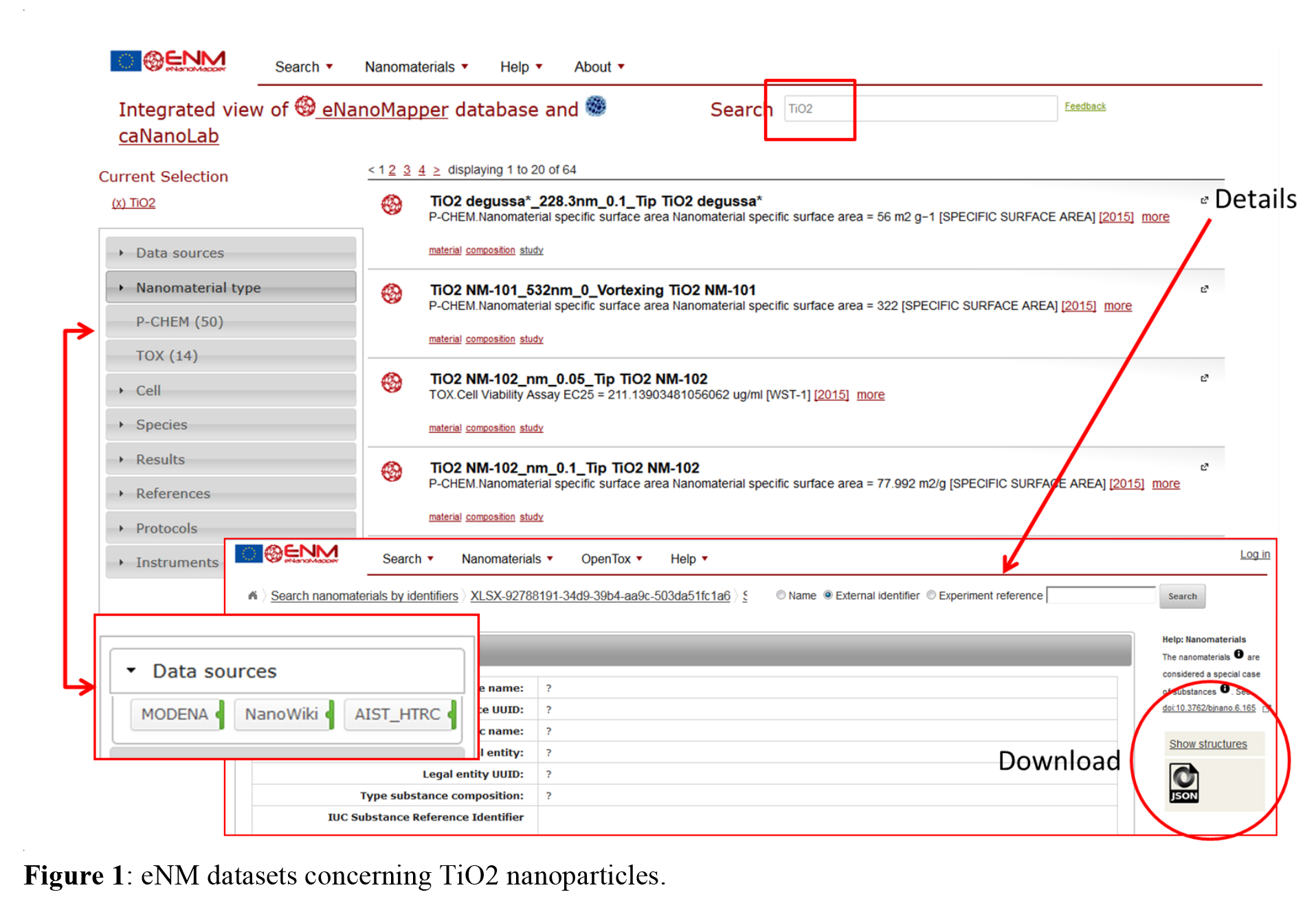
Specific nanoparticles can be found by using either nanoparticle name, identifier, composition, or free text search. Entering e.g. TiO2 in the “name search” will give the list in figure 1 of available data. The grey column on the right side allow filtering for different terms like data sources, nanomaterial types and others. Details for the single studies can be found by clicking on the links and the derived results can be downloaded in form of JSON, CSV or RDF files for further application.
2. How to find data of carcinogenicity experiments with nanoparticles?
The eNM database offers the possibility to search specific data containing phys-chem or biological assays (e.g. Search - Search nanomaterials by physchem parameters or biological effects) figure 2. Then, select the assays in question on the checklist at the left side (e.g. Carcinogenicity assay) and apply “update results”. The resulting list will show all database entries with that assay and provide a possibility for download (figure 3).;
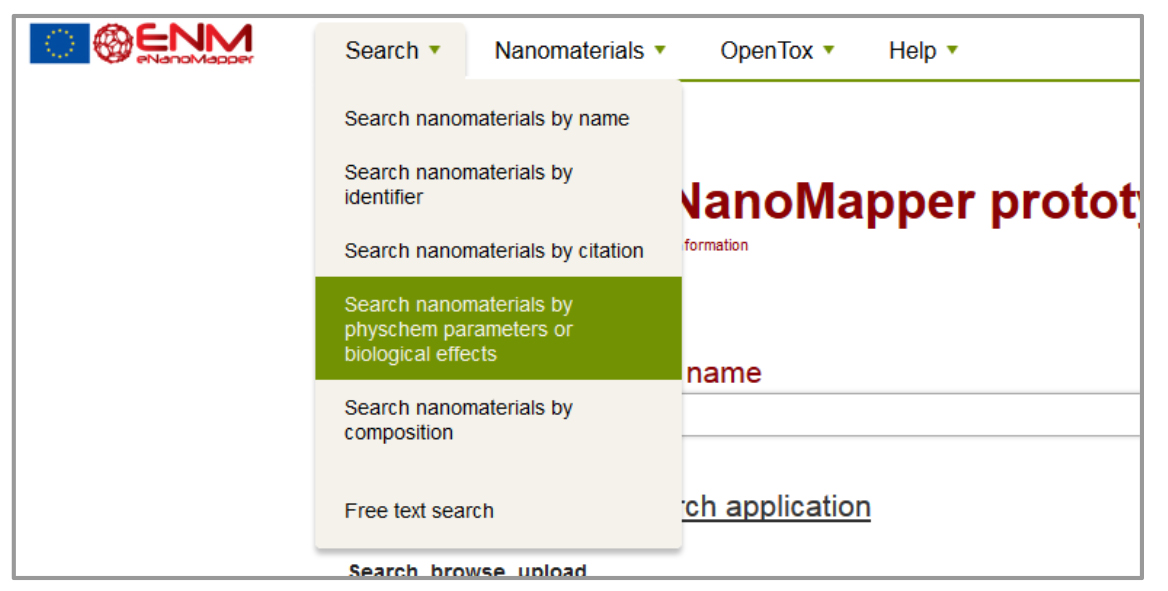
Figure 2: Search function.
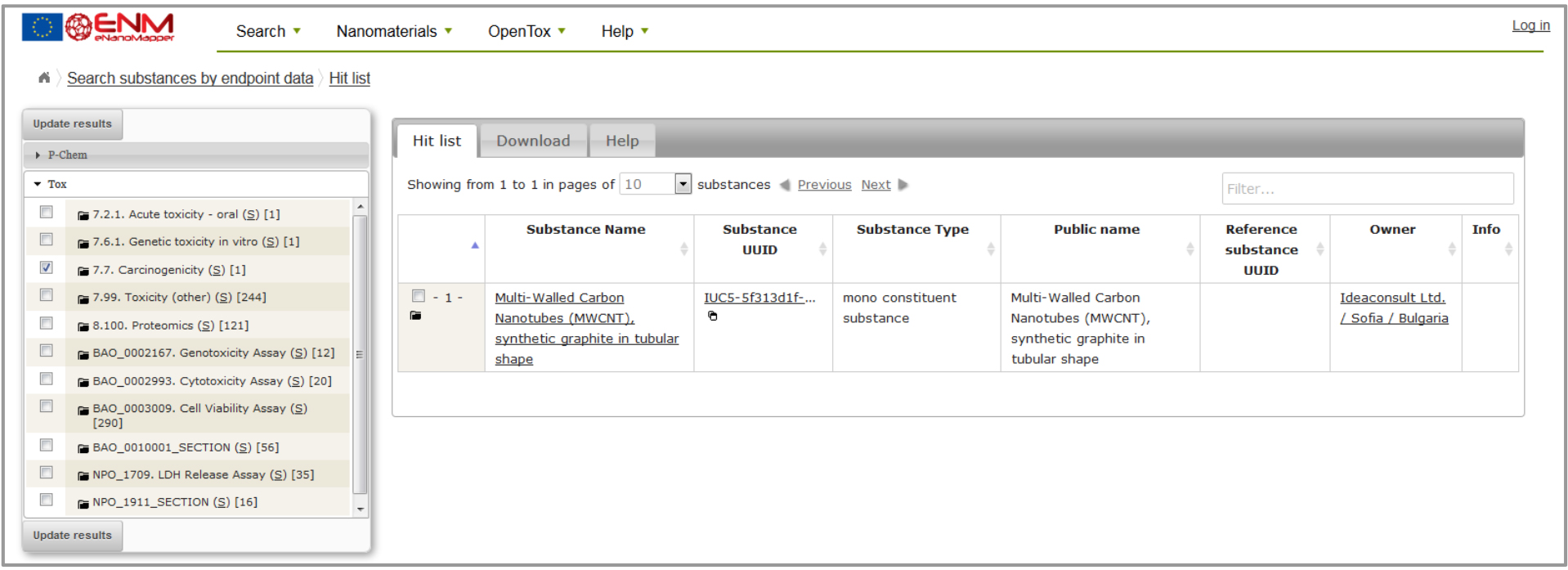
Figure 3: Result example in the eNM database for carcinogenicity data.
3. How can we merge data from different assays?
Using ontology terms allows to merge several assays (including similar experiments). The physchem/biological effect search (see figure 2) allows to filter the results for general terms like cell viability or cytotoxicity which include a variety of different assays. The list is dynamic and the number and the specificity of the assays in the list depends on how the imported data was annotated. For example, if asked for datasets with cell viability endpoints, the current database returns 80 datasets. Searching specifically for LDH assay (lactate dehydrogenase release assay, an indicator for membrane integrity and therefore cell viability) there are 17 datasets from FP7 MARINA and MODENA projects. With more (properly annotated) data being imported the list will extend. The filter functions in the hit list are currently limited to the textual information available in the table (i.e. substance names and study owners, external identifiers). Desirable would be also terms which allow filtering and ranking for quality measures like data size, data completeness, and date of data production (or publication).
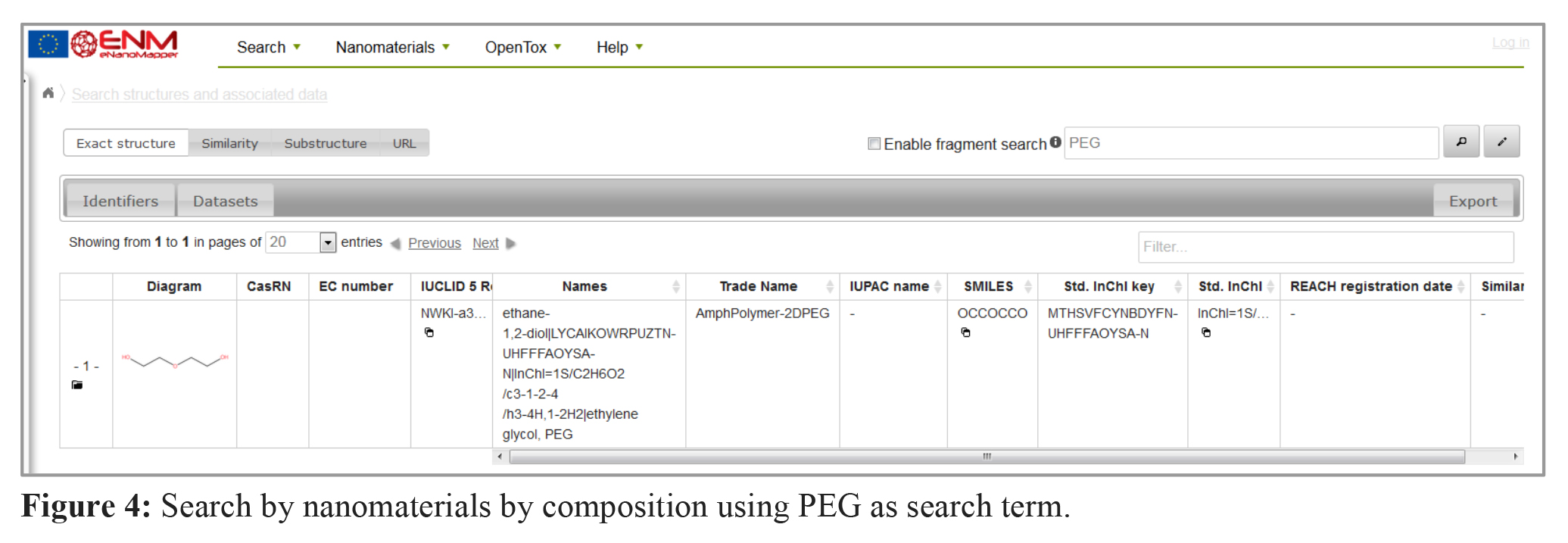
4. Which nanomaterials in the database contain PEG?
To get data of nanomaterials containing a certain material in core, shell or linker material, the search function “nanomaterials by composition” can be used. The hits are labelled (e.g. core or shell). The example in figure 4 shows the currently available data about PEG (polyethylene glycol) containing nanoparticles in the eNM database.
5. What happens if there is nothing in the database about my material?
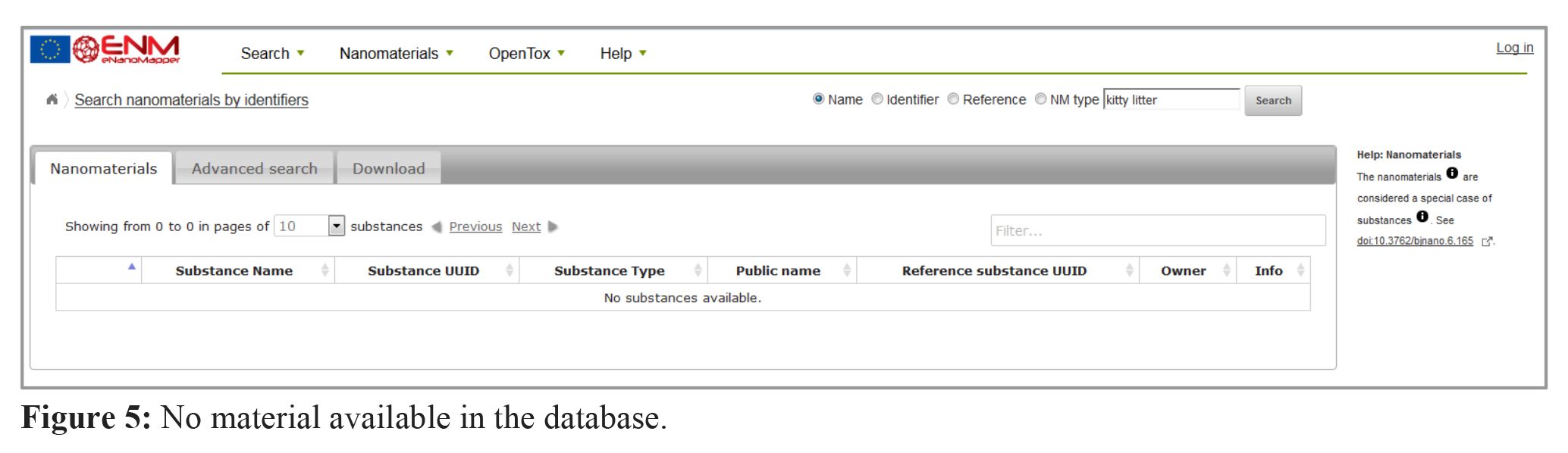
Looking for material which is not yet in the database (e.g. “kitty litter”) will return “no substances available” as shown in figure 5. Please feel free to upload your data (see this tutorial, 3. Data preparation and upload) or contact the eNM consortium.